Early this morning Marty McFly has arrived to the future. It is true that cars don’t fly quite yet. Hoverboards are not available either. Households are not powered by their own nuclear reactors and clothes don’t dry and adjust automatically. Good old Marty has got reasons to be disappointed. But advances in some other directions have been astonishing during the last 30 years. Who could foresee the transformative power of the internet? Everyone is plugged to a tablet or smartphone, immediately accessing far away friends and personalized web content. Synthetic biology is taking its first, promising steps (to which we are glad to contribute) and advances in prosthetics and Brain Computer Interfaces are bringing back motility and communication to injured patients.
An early breakthrough in Brain Computer Interfaces (BCIs) was the P-300 speller. These devices show a matrix of letters with rows and columns that are highlighted in a random order. The user of the interface focuses on the position of the matrix containing the letter that she wishes to spell. Whenever the letter is highlighted, a very characteristic signal called P-300 mounts up in her brain and is readily picked up by an electroencephalogram (EEG) apparatus. This way, people without motility can build words and sentences, literally, right out of their thoughts.
P-300 is a complex macroscopic signal of our brain. Its most robust feature is a positive deflection of the electric potential that can be registered in the central channels of the EEG. This signal is associated to perceptions (often visual or auditory) that catch your attention over other, more neutral stimuli. Nice examples are your next letter being highlighted above others in the matrix speller; a random, high-pitched tone amid a regular series of low notes; and, arguably, someone calling your name in the middle of a crowd. Some people suffer terrible conditions that do not allow their gaze to roam freely, so they cannot focus on distinct positions of the matrix. Rapid Serial Visual Presentation (RSVP) of letters right at a user’s fixation point allow them to benefit from fast BCI spellers anyway.
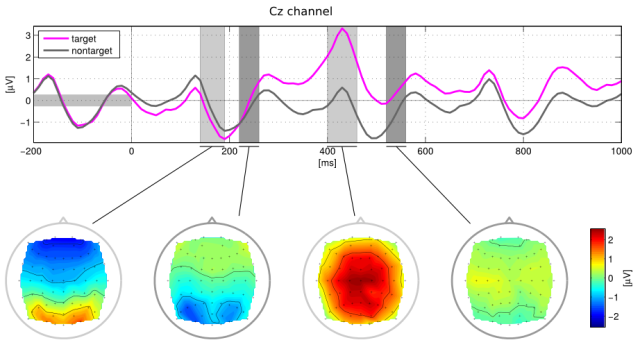
Above: Average electric activity at the central channel of an EEG reproducing P-300 (violet) and background (gray) signals. Below: Average electric activity in a scalp during P-300 activity.
And why should we stop with letters? RSVP and P-300 make the perfect scenario for sci-fi movies but also for serious research and technological developments. For example, we could throw more or less random polygons, filled with different colors, at a blank screen and hope that some of them will catch the attention of a user connected to an EEG. Could these polygons elicit a P-300? Perhaps, if they remind the user of some image in her mind… if they match a tiny detail or the overall color of that scene she’s thinking about! We could proceed incrementally, capturing these tiny details and accumulating polygons to form more complex drawings — just as we do with letters in a sentence.
This idea was envisioned and developed by Complex Systems Lab member Luís Seoane and his colleagues Benjamin Blankerz, PI at the Berlin Brain-Computer Interface, and Stephan Gabler then at the Bernstein Center for Computational Neuroscience in Berlin. In a paper published this summer they show how those Images from the Mind could be reconstructed based on the P-300 EEG signal. While the images where fairly simple and the settings of the experiment were cautious, the very positive results indicate that faster BCIs for image reconstruction are possible. Some interesting future designs are discussed in the paper as well. We hope that this work makes a nice contribution to this relevant technology that can greatly impact the lives of many people.
Back to Marty and Doc’s adventures for today, surely BCIs can help them finish their chess game against Einstein. We hope that they are enjoying the unexpected twists of technological evolution that brought us this far… at least while all the other promised gadgets don’t get going.
Some relevant links from the text:
- Video of a P300 speller by the Neuronal oscillations and cognition group, Dept. Intergrative Neuophysiology, CNCR, VU University Amsterdam.
- Paper by Laura Acqualagna and Benjamin Blankertz proposing a RSVP speller.
- Seoane, Gabler, and Blankertz’s paper proposing a RSVP-based BCI to reconstruct images with the mind.
- Successful reconstruction of a cherry.






 ANGEL RAYA, Fronteres de la manipulació genètica humana
ANGEL RAYA, Fronteres de la manipulació genètica humana MAVI SANCHEZ-VIVES, Un cervell molt plàstic: transformacions corporals en realitat virtual
MAVI SANCHEZ-VIVES, Un cervell molt plàstic: transformacions corporals en realitat virtual